Guideline for AD-LitPathoNet Network Services

AD-LitPathoNet is an abbr. for “Alzheimer’s Disease – Pathology Network with Rich Literature
Evidence“.
Visit us: http://lit-evi.hzau.edu.cn/AD-LitPathoNet/
The AD-LitPathoNet database is a user-friendly AD pathology investigation platform that enables investigation of local-to-global pathology mechanisms, macro-to-micro pathology network visualization, and evidence tracing. The database data is automatically collected from approximately 300,000 PubMed abstracts and 90,000 PMC full-text articles related to AD using BioNLP-based methods, and the data source is updated quarterly.
1. Knowledge Representation Paradigm of Pathology Mechanism for AD
A “Traceable pathology mechanism representation” is set up for the knowledge representation paradigm
in support of AD pathology mechanism.
The biomarkers used for the pathology illustration include gene and mutation, while the phenotypes
descriptors include GO, HPO and MeSh.
First, mutation, gene (or protein) and the “occurrs_in” relation is mined from the texts. Subsequently, the
downstream biological process, standardized by GO
terminology, is retrieved. Meanwhile, the “causes” relation between a mutation and a downstream biological
process is recognized via intelligent NLP method. By doing so, the “traceable pathology mechanism
representation” captures a clear pathology description that a mutation “occurrs_in” a gene “causes” a
downstream biological process. As well, this knowledge representation is traceable since the supportive
literature evidence is stored simultaneously
In summary, the “traceability” of AD-LitPathoNet refers to two respective.
▩ First, the logic of biomarker and downstream phenotype is accurate and traceable. The causal relationship between biomarker and the downstream phenotype forms the basis for pathology mechanism illustration in the AD context.
▩ Second, the sentence-level literature evidence is traceable.
Based on large-scale text mining, knowledge curation, and evidence tracing, AD-LitPathoNet makes it possible to explore the AD pathology mechanism and trace the evidence systematically.
2. Data Service Modules in AD-LitPathoNet
AD-LitPathoNet provides three main modules:
"Auto-Completion Query", "Go Visualization", and "Systematic Analysis".
▩ "Auto-Completion Search" for Phenotype or Genotype query.
The "Auto-Completion Search" module provides a user-friendly dropdown selection feature for phenotype and gene searches.
▩ "Go visualization" for Phenotype or Genotype query.
The "Go Visualization" module provides interactive visualization of macro-to-micro pathology networks exploration and literature evidence tracing.
▩ "Systematic Analysis" for Phenotype or Genotype query.
The "Systematic Analysis" module includes the "Local Association Recommendation" and "Global Interaction Enrichment" sub-modules to enable the investigation of AD pathology mechanisms at both local and global levels.
3. Get started with a query
AD-LitPathoNet provides two types of data services for both phenotye and gene query.
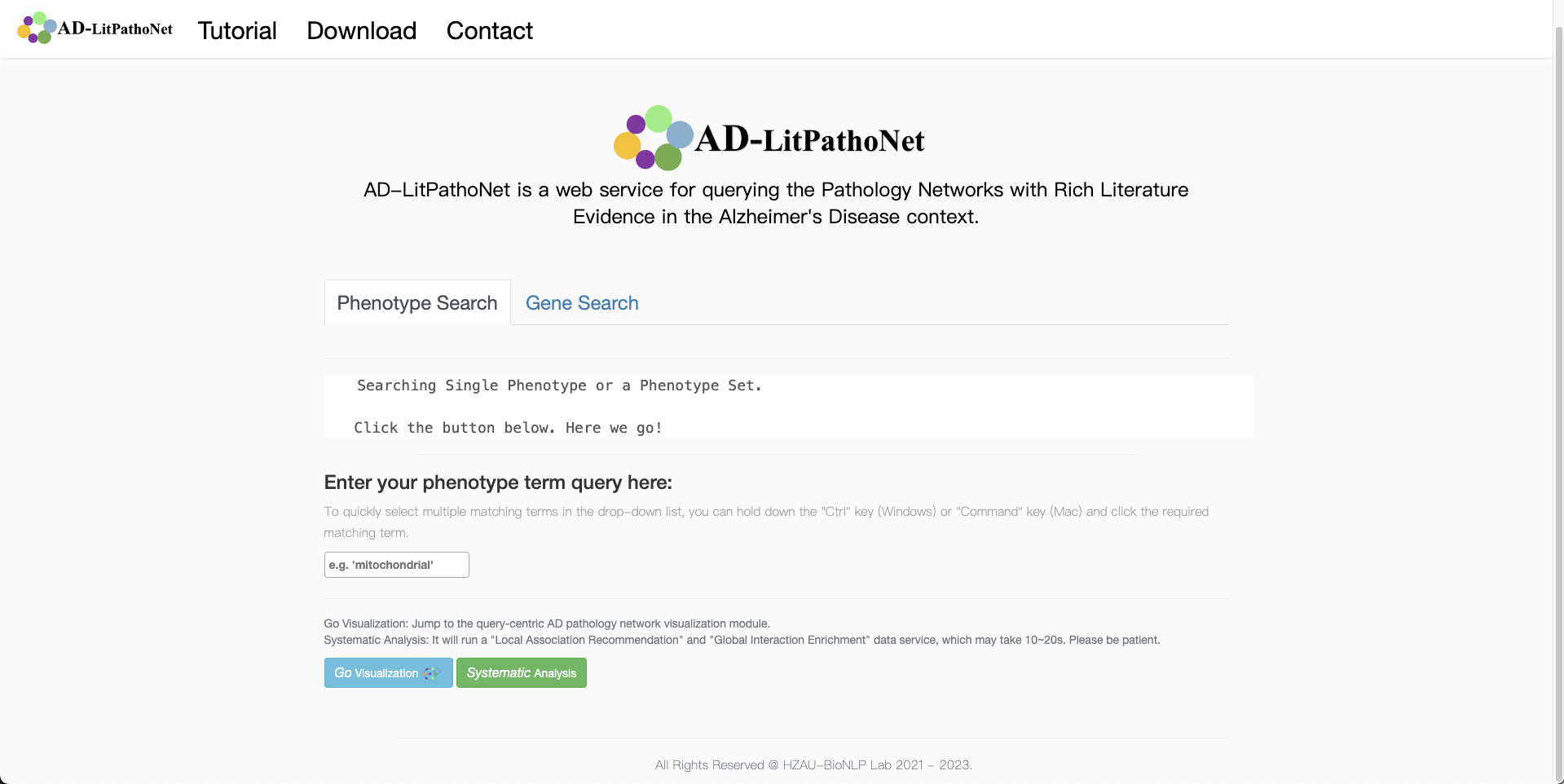
- In the default tab for the main page, "Phenotype Search", an auto-completion drop-down box is adopted, allowing users to select single or multiple terms of interest. AD-LitPathoNet will provide all the matched terms in the database for selecting.
- Alternatively, the user can click on the "Gene Search" tab to submit a single gene or gene set by name, with the same search pattern as the former.
- Users need simply press Enter or click the "Go Visualization" button to be taken to the pathology network visualization page. And click the "Systematic Analysis" Button to obtain the summary report which contains the association recommendation and interaction enrichment modules.
▩ Basic phenotype and gene query submission examples
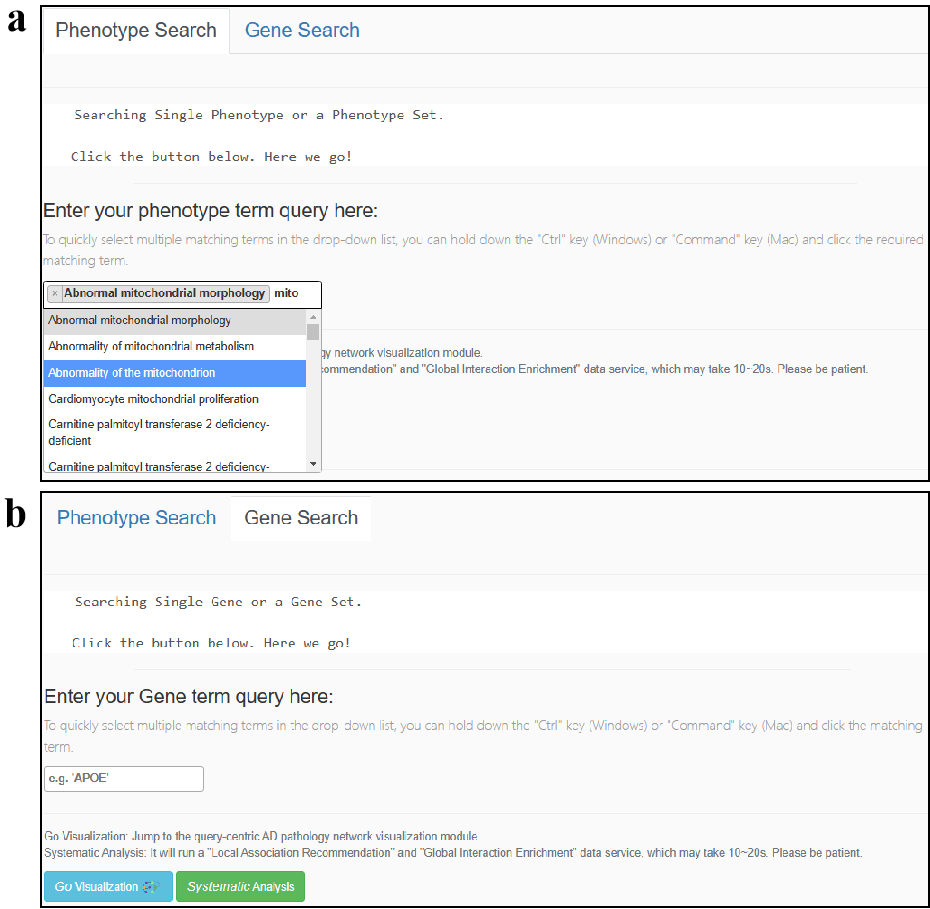
4. "Go Visualization": Obtain the visualization page
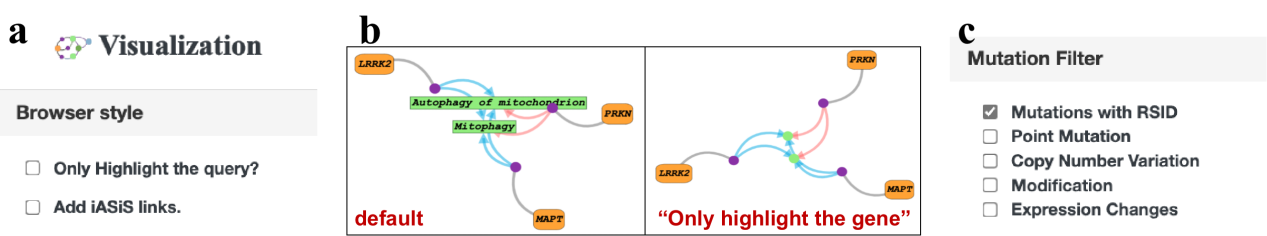
The visualization in AD-LitPathoNet provides user-friendly data investigation in terms of pathology tracing. A "browser style" option is provided to customize the appearance of the network diagram by highlighting particular queried terms. When the user checks "Only highlight the gene", the phenotype terms become a dot and does not show the full name . iASiS is our collaborative database, so if users want to get more "phenotype-phenotype" interactions, they can check the "Add iASiS links". By restricting mutation type options, users are able to narrow down the search and pinpoint the interested knowledge term. Five types of mutation options are provided, i.e., RSID normalized mutations, point mutations, copy number variants (CNV), modifications, and expression changes, and only the mutations with RSID is selected by default.
▩ Network filter box
In the network visualization area, the user can filter the node type by the node labels at the top, and the network diagram download, network layout refresh and full screen display are provided in the upper right corner. The "occurrs_in" relation between a mutation and a gene is represented by a grey line. Meanwhile, the "causes" relation between a bio-marker and a downstream phenotype is visualized by a red, blue, or black arrow, which defines positive regulation, negative regulation, and association, respectively. Literature evidence and external resource for each node and edge are presented, which allows users to explore the supporting evidence for each connection and jump to external resources for further information. Users just need to click on the node or edge of interest to see the literature evidence and external resource.
▩ The network of "Mitochondrial fission"
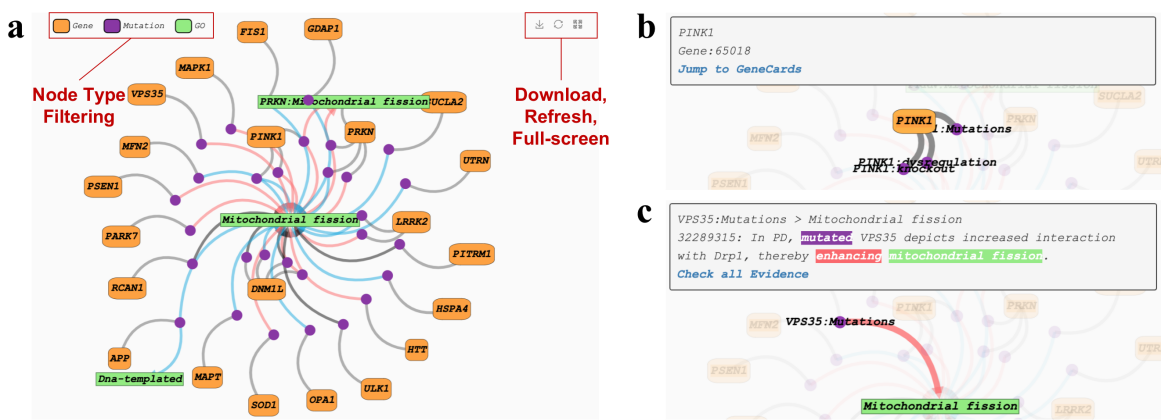
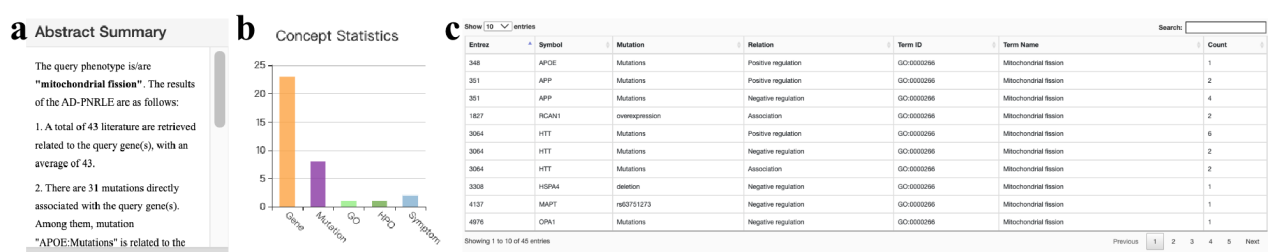
In the network and evidence presentation, uniform color annotation is used for each entity (genes are orange, mutations are purple, GO is light green, HPO is dark green, MeSH is blue) In particular, AD-LitPathoNet provide abstract summary, statistics on the number of nodes , and a tabular presentation of the corresponding visual network on the same page.
▩ Additional information on the network of "Mitochondrial fission". a. Abstract summary describes the literature involved in the network. b. Concepts statistics is a histogram on the number of nodes. c. Table is a tabular presentation of the interactions in the network, with each row representing a gene (Symbol) affecting (Regulation) a phenotype (Term) through a mutation (Mutation).
5. "Systematic Analysis": Discover more
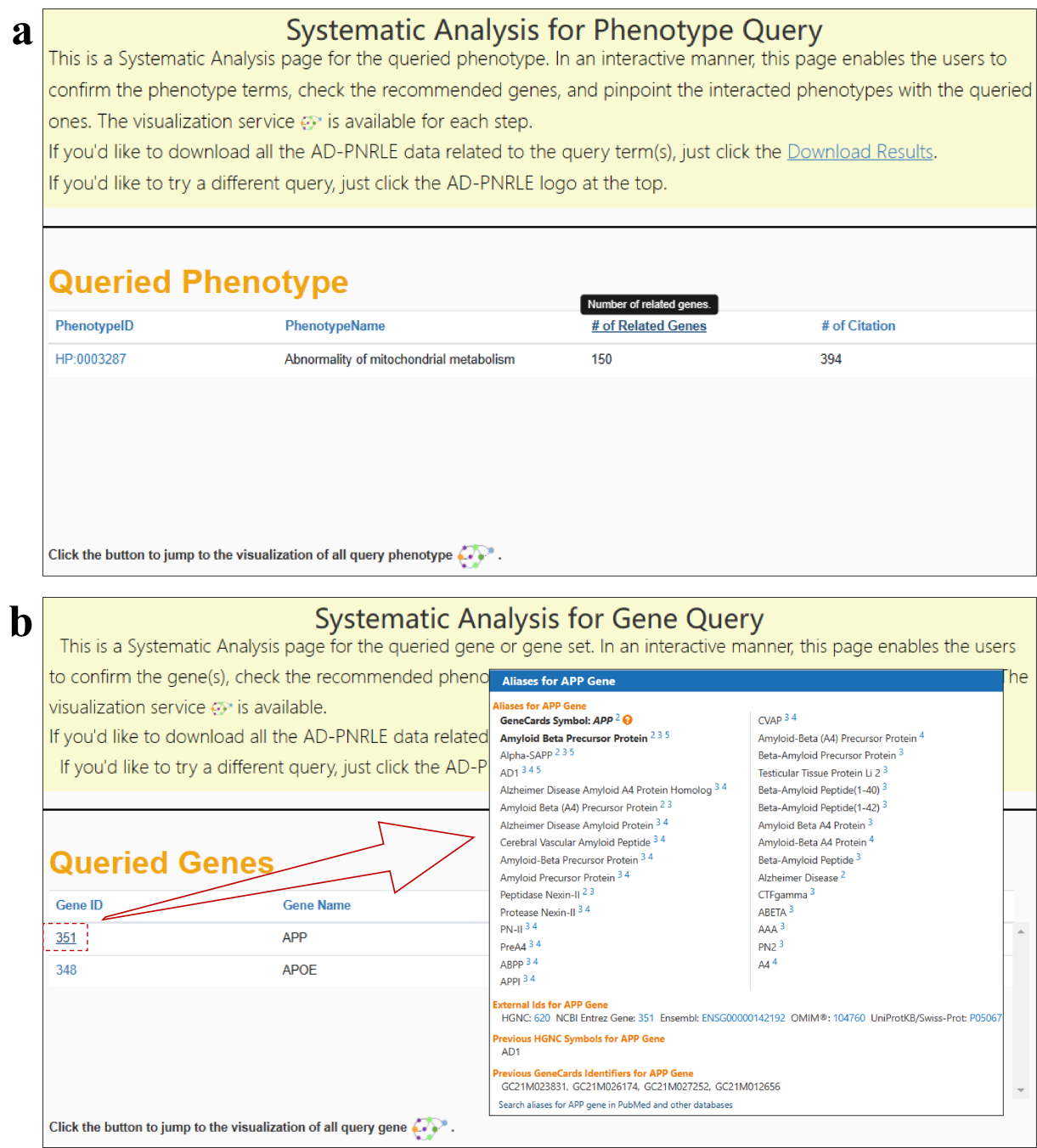
In the result page of Systematic Analysis, the user is first presented
with a brief description of the page, and a link to download the data (click the "Download Results"). In
each of the data service modules in AD-LitPathoNet, there are hoverable tips with detailed information about
the headers of each data table and what it means. Some data have hover-tips within the tables
as well, such as the PhenotypeID. Some data are supplemented with external links, i.e. clicking on a Gene ID
will jump to the corresponding GeneCards page.
The first module shows the statistics of the query terms in the AD-LitPathoNet
database, including the
number of related gene (for Phenotype Search) or phenotype (for Gene Search) and the number of citation.
At the bottom of the module, there is also a link to a visualization for the
query term, where the
visualization is the same as the results obtained by clicking the "Go Visualization" button on the home
page.
▩ Navigation of Systematic Analysis. (a) Result of querying "Abnormality of mitochondrial metabolism" matched by "Phenotype Search". (b) Result of querying "APP APOE" matched by "Gene Search". Click on a gene ID will jump to the corresponding page of GeneCards.
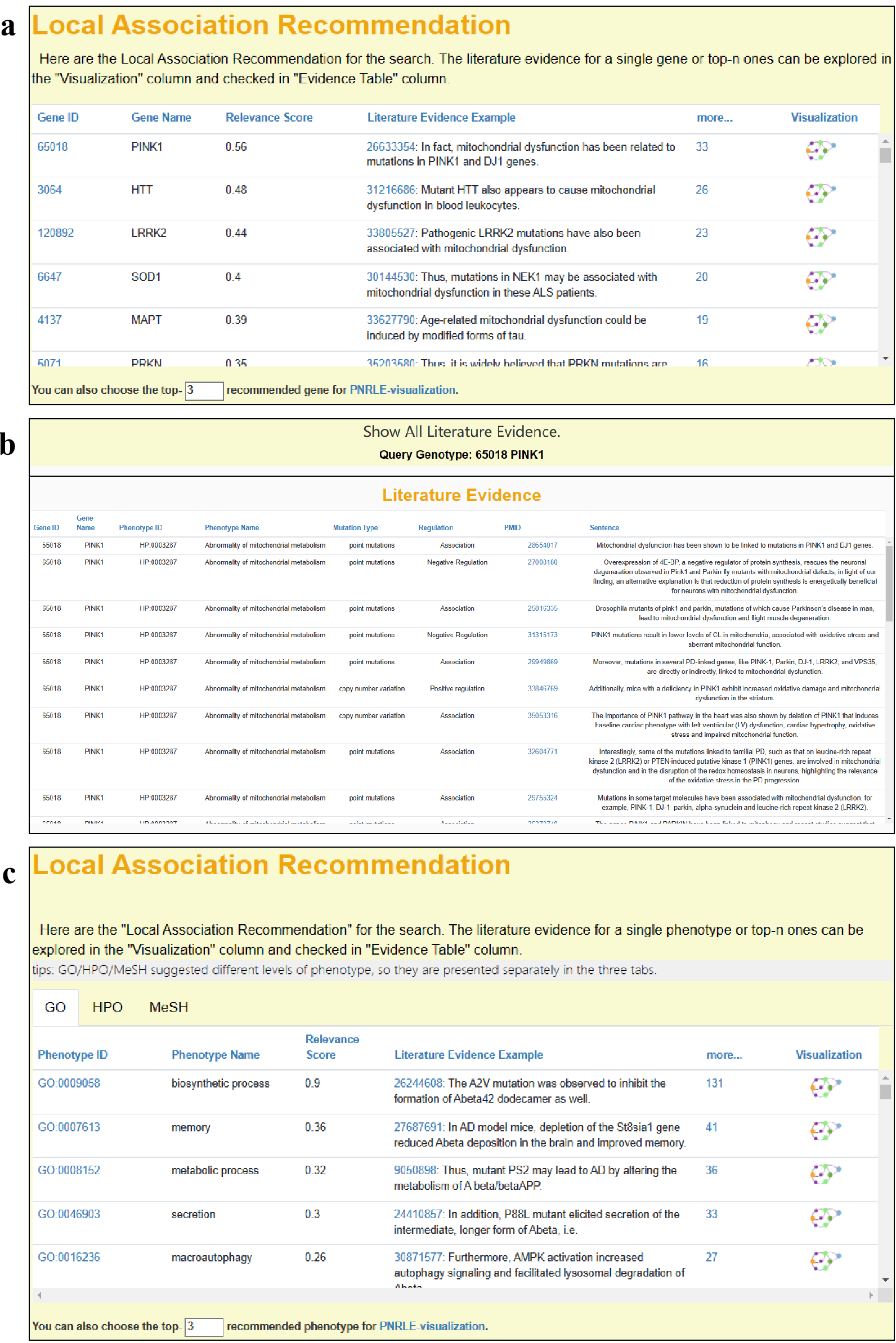
5.1 "Local Association Recommendation"
In this module, a TF-IDF score-based recommendation system is used
by the AD-LitPathoNet to generate a list of genes or phenotypes, these terms are extracted from
literature associated with the user's query.
The association recommendations for both searches are presented in a similar
tabular format, with relevance score, literature evidence example, a full evidence table and evidence visualization for each term.
Click on the PMID in the Literature column to jumps to the PubMed of source literature.
▩ Local Association Recommendation Module. a. Association Recommendation result of Phenotype Search for "Abnormality of mitochondrial metabolism". b. All literature evidence table of PINK1 which is the top-1 gene in Figure. c. Association Recommendation result of Gene Search for "APP APOE".
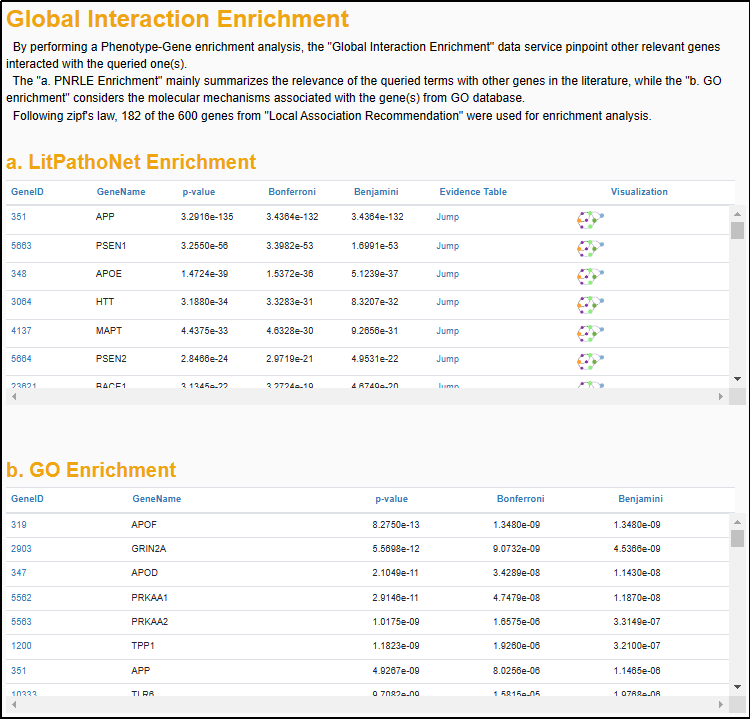
5.2 "Global Interaction Enrichment"
The association data in AD-LitPathoNet enables an interaction enrichment analysis that investigates the phenotype-phenotype interaction in a specific phenotype context, or the gene-gene interaction in a specific gene(s) context.
▩ Interaction Enrichment results for the Phenotype Search query "Abnormality of mitochondrial metabolism".
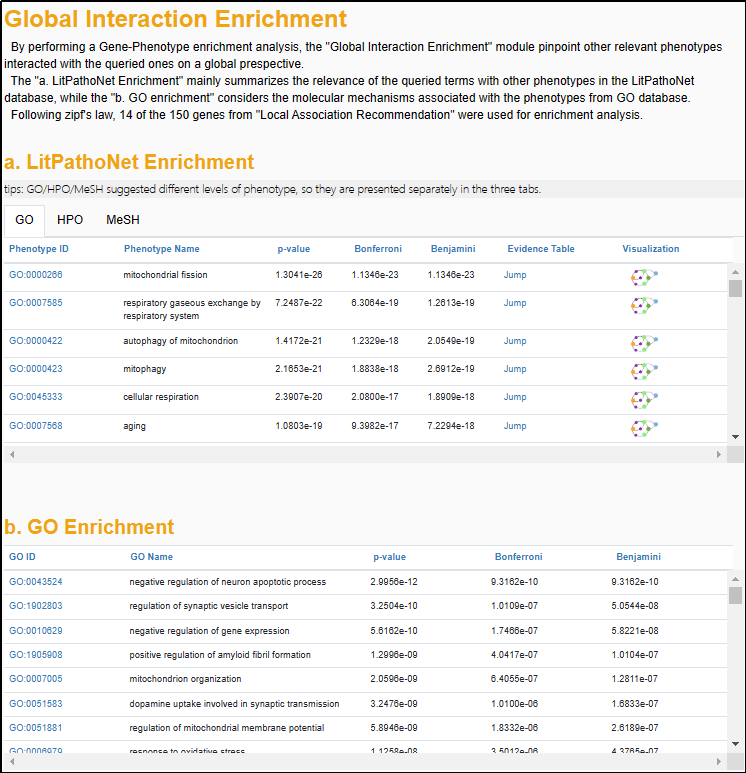
To achieve more accurate enrichment results, following Zipf's law, only terms that accumulate a score proportion of over 80% from the previous step are used for further analysis. Then, AD-LitPathoNet took LitPathoNet database and GO as background dataset respectively and calculated P-values by using hyper-geometric distribution for genes or phenotypes obtained from association recommendation module. Both Bonferroni method and Benjamini method were used for P-value correction.
For instance, in the Phenotype Search, Association Recommendation module provide a couple of gene related to the specific phenotype query. Then, through gene-phenotype enrichment analysis, AD-LitPathoNet in turn suggests phenotype terms enriched for these genes. Similarly, in the Gene Search, the gene-gene network mediating common phenotypes is obtained by phenotype-gene enrichment analysis. For data in the context of the LitPathoNet database, full Evidence Table and Visualization are provided for users to investigate further.
▩ Interaction Enrichment results for the Gene Search query "APP APOE".
The association recommendations for Gene Search are presented as tabs with three phenotype descriptions (GO, HPO and MeSH). At the bottom of this module, users can enter the number n and then click on the link at the end of the sentence to see the visualization of the association network among the query term and Top-n phenotypes/genes.
6. A PROOF-OF-CONCEPT EXAMPLE FOR GENE SEARCH
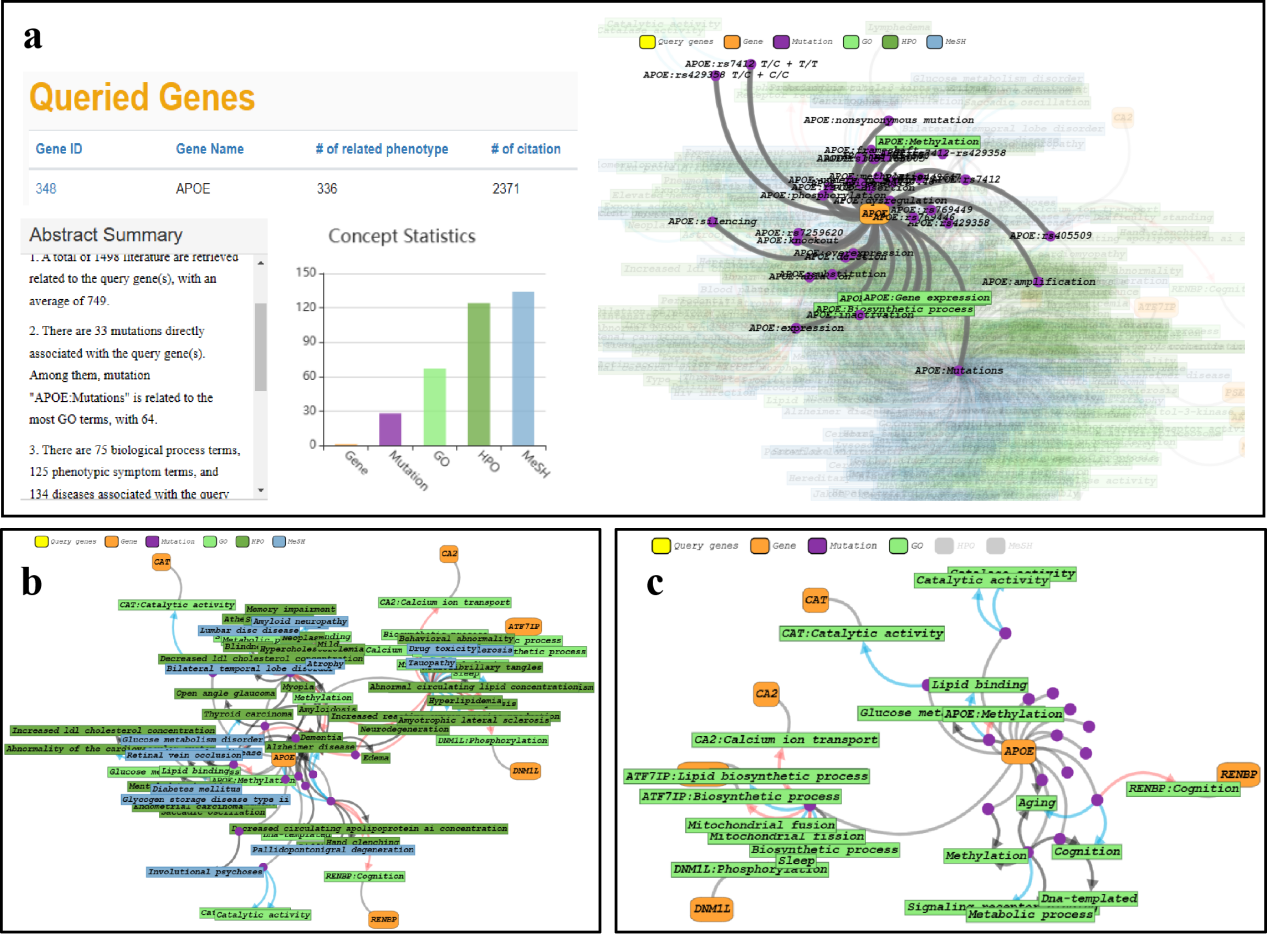
5.1 Overview of holistic situation
Taking the gene APOE as a query example, which is the major genetic
factor in late-onset Alzheimer's disease, thus, a better organization of the literature evidence on APOE
in AD research could help illuminate the mechanisms of AD pathology.
According to the statistics, APOE is cited in 1,498 literatures in the
AD-LitPathoNet database, associated to 325 phenotypes (67 GO, 124 HPO, 134 MeSH). The visualization
network is quite complex for such a large amount of data search terms, so it is possible
to select the mutation types of interest by the filter box on the left, or to hide some of
the node types by the labels above the network. Alternatively, in the subsequent
calculation module, the pathology visualization network between the search terms and the recommended
biomarkers can be viewed in a targeted manner.
▩ Overview of the holistic situation of APOE in AD-LitPathoNet. a. Statistical information and visualization of the terms associated with APOE in the database. b. Visualization network that only shows mutations with RSID on the basis of Figure. c. Visualization network after hiding HPO and GO entries on the basis of Figure.
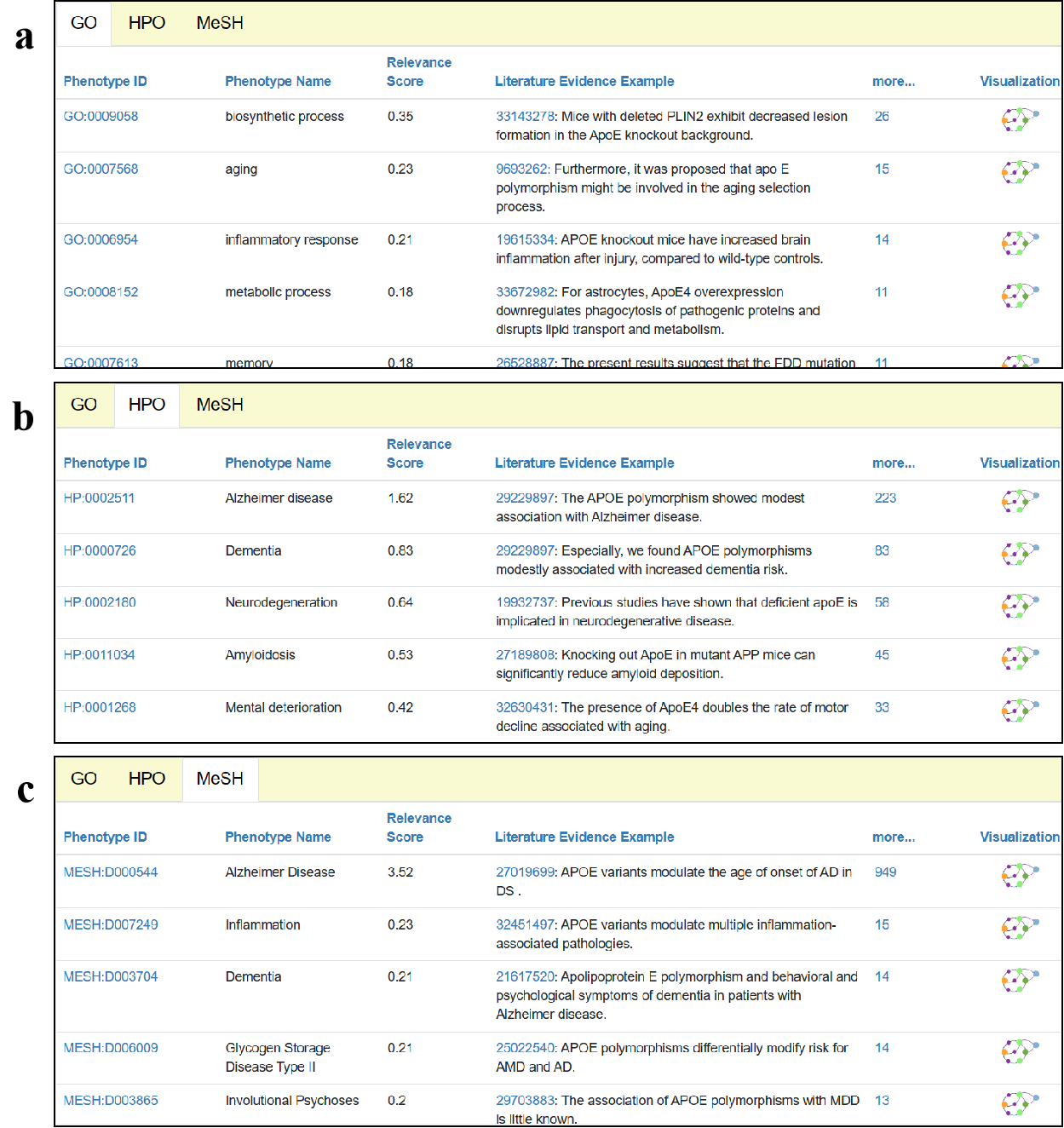
5.2 Knowledge discovery from the local to global
The Association Recommendation module of AD-LitPathoNet allows users to trace the most related phenotypes about APOE and explore the gene-phenotypes associations. This module suggests 336 associated phenotypic terms, where the higher scoring terms mostly belong to HPO and MeSH, including "Alzheimer disease", "Dementia", "Neurodegeneration", and obviously, these terms are all related to AD. For the GO terms, the main biological processes involved in APOE are listed, such as "biosynthetic process", "aging", "inflammatory response", etc. All of these phenotypes are specifically associated with APOE and together form its local knowledge network.
▩ Results of Association Recommendation for APOE. a. Recommended GO terms. b. Recommended HPO terms. c. Recommended MeSH terms.
Based on the above phenotypes, Interaction Enrichment are performed for the top 76 related phenotypes according to Zipf's law. In the LitPathoNet background, a group of genes that shared common phenotypes with APOE are obtained, such as PSEN1, APP, HTT. And the enrichment in the GO background complements the above results. Interaction Enrichment modules link the single gene of "APOE" to other genes via a set of recommended phenotypes in the setting of AD, thus expanding the knowledge network from local to global.
▩ Results of Interaction Enrichment for APOE.

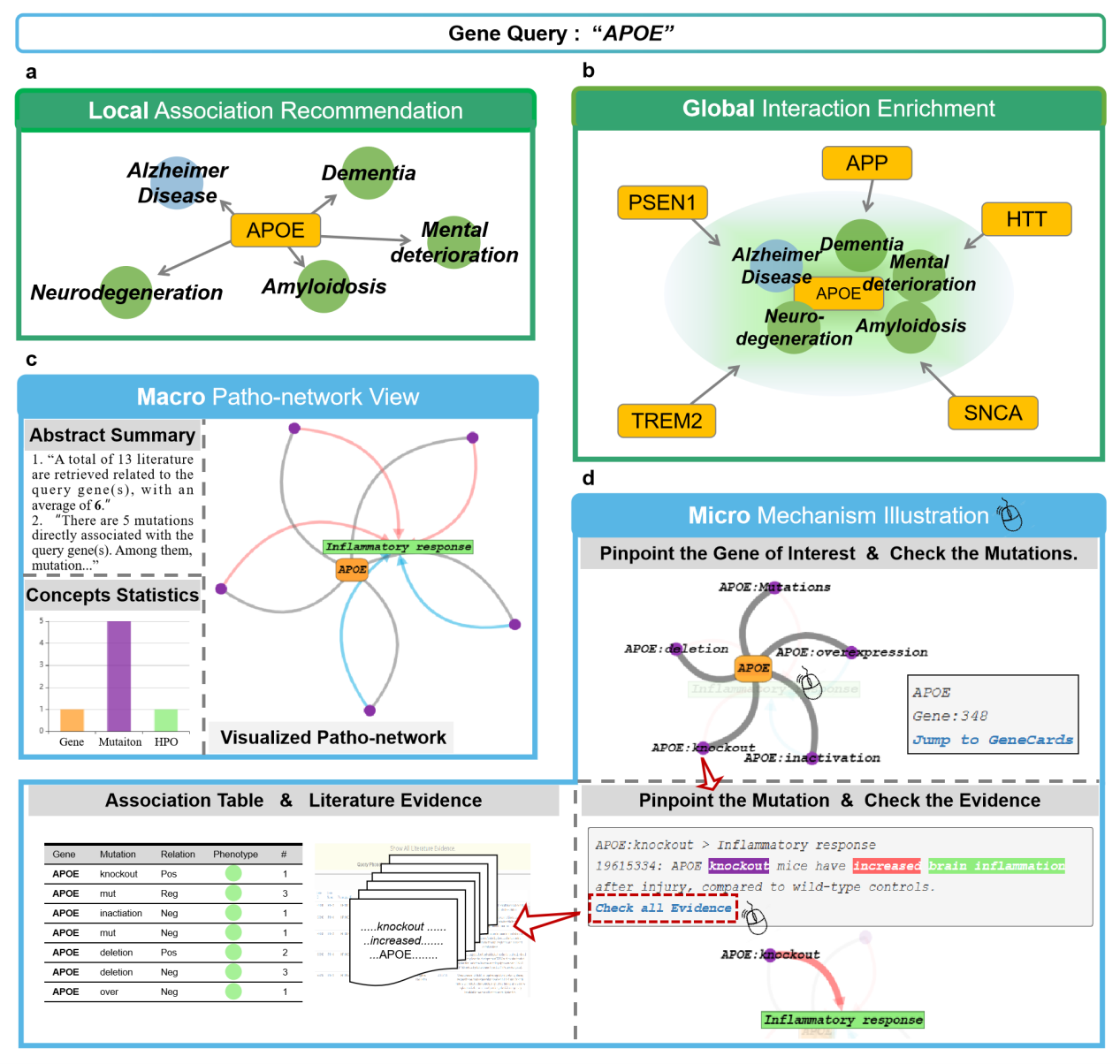
5.3 Pathological exploration from macro to micro
To explore the pathological mechanisms of AD in a certain setting, pathology visualization networks for single/multiple genes or phenotypes can be viewed at all of the above data services. We select the association between APOE and its recommended phenotype "inflammatory response" to illustrate the macro-to-micro pathology discovery. Under a macro view, by integrating abstract summary, concept statistics, and visualization network, all data related to the association involved 13 publications containing 5 mutations ("deletion", "knockout", "inactivation", "overexpression" and "Mutations"). Further, in a microscopic view, we can choose to check the literature evidence with which a biological process is reported. Thus pathology knowledge of a causal mutation and a regulated phenotype is derived.
▩ Gene Query. An example query using "APOE" as the searched term. a. Association Recommendation. This module provides a set of phenotypes that are specifically associated with the query term. For gene "APOE", the top five recommended phenotypes are "Alzheimer disease", "Dementia", "Neurodegeneration", "Amyloidosis", "Mental deterioration". b. Interaction Enrichment. Based on the top 76 phenotypes obtained in the previous step, a group of genes e.g., "PSEN1", "APP", "HTT" and so on, are collected by enrichment analysis, and it can be concluded that "APOE" is associated with these genes. c. Macro Patho-network View. Abstract summary, concepts statistics and visualized patho-network together form a macroscopic view of the relationship between "APOE" and "inflammatory response". d. Micro Mechanism Illustration. In addition, the users are able to obtain and download the interested data investigation result in the forms of association table and full literature evidence report.
ENJOY!